We're following the overview tuturial (here), which has four main parts. There are the three big sections of
custom_parameters.txt: plus
Today we're thinking about alpha diversity (within sample diversity). Goals of diversity analysis (using 16S rRNA sequences in bacteria) include measurement of:
where OTUs can be defined at different thresholds of sequence identity.
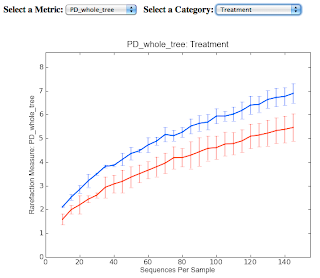
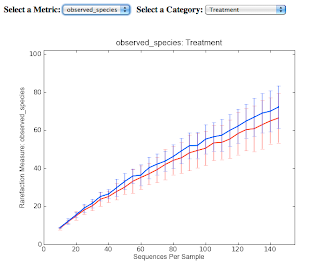
For this post, I've got screenshots of two graphics produced from the Qiime tutorial analysis of alpha diversity. Consider the first graphic, which plots observed species in two different samples as a function of the number of individual sequences examined. Because chance influences the order in which samples are obtained, resampling techniques are used to generate a rarefaction curve that averages the results of many random samplings of the observed data. Perhaps more important, it allows normalization for the number of samples observed, allowing comparison of samples with different sizes.
These plots are rarefaction curves.
The most important questions are probably these two:
(1) If we could sample exhaustively, what would be the final species count (or phylogenetic distribution or.. ). In other words, does the curve level off, and what is the asymptotic value?
(2) Can we decide whether two different populations differ significantly even without exhaustive sampling?
I do not know much about this area, so I should probably just be quiet at this point, but I have to say that I am suspicious that the first question does not always have a good answer (even if people would wish for one). The problem is that the shape of the curve as it goes out into high number of samples does not necessarily depend on the shape at lower numbers. It might do so, if the population structure is not too skewed. But no one can say in advance whether that is true or not.
Anyway, we follow the tutorial. First remake (and this time, save) the otu_table:
We'll use the workflow script (I don't see too much of interest after a cursory look at the intermediate steps). We can get the required options with -h (help):
What we actually run is:
command:
The default settings in
custom_parameters.txt are fine for this.If you do want to run each of the steps individually, I suggest you run the workflow script, then the log file in will contain a history of the commands that were executed:
The second graphic shows that considering phylogenetic diversity may reveal significant differences that are not seen when just counting OTUs.