The FASTA format is defined here. There are various problems one can run into with long titles, including the presence of spaces, character limits, and a requirement for unique names within those limits. For now, I'll just remove most of the title, leaving '>' plus the Genbank id from the end of the line. (If the identifiers are already known, I have a Python script that will retrieve the sequences from Genbank automatically). Now I have 5 sequences saved in 'strep.txt':
X58303 Streptococcus mutans
AF003928 Streptococcus sanguinis
AF003930 Streptococcus pneumoniae
AB002521 Streptococcus pyogenes
AF003933 Streptococcus parasanguinis
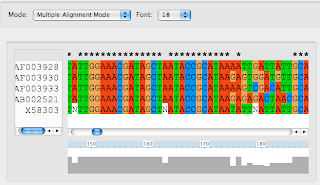
Next I run clustal to get a sequence alignment. I got clustalx and clustalw from here. (Click on Download Software). Load the sequences into ClustalX (2.0.5). Under Alignment => Output Format Options select 'FASTA format' and then select 'Do Complete Alignment.' It looks like this:
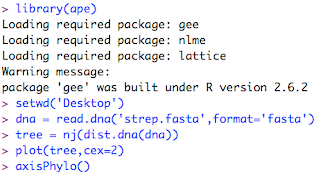
Now, to plot the tree. ClustalX has given us an output file named 'strep.fasta.' I start up R (get it here). I have previously used the Package Manager to install a phylogenetics package called 'ape'. For more documentation you can search the R web site for 'ape'. This is my interaction with the R interpreter:
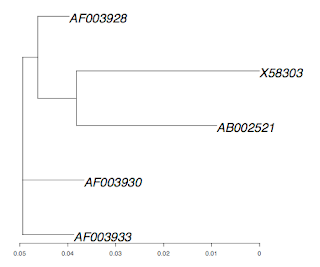
And here is the plot of the tree: